graphisomorphism
Find isomorphism between two graphs
Syntax
[Isomorphic, Map]
= graphisomorphism(G1, G2)
[Isomorphic, Map]
= graphisomorphism(G1, G2,'Directed', DirectedValue)
Arguments
G1 | N-by-N sparse matrix that represents a directed or undirected
graph. Nonzero entries in matrix G1 indicate
the presence of an edge. |
G2 | N-by-N sparse matrix that represents a directed or undirected
graph. G2 must be the same (directed or
undirected) as G1. |
DirectedValue | Property that indicates whether the graphs are directed or
undirected. Enter false when both G1 and G2 are
undirected graphs. In this case, the upper triangles of the sparse
matrices G1 and G2 are
ignored. Default is true, meaning that both graphs
are directed. |
Description
Tip
For introductory information on graph theory functions, see Graph Theory Functions.
[
returns logical 1 (Isomorphic, Map]
= graphisomorphism(G1, G2)true) in Isomorphic if G1 and G2 are
isomorphic graphs, and logical 0 (false) otherwise.
A graph isomorphism is a 1-to-1 mapping of the nodes in the graph G1 and
the nodes in the graph G2 such that adjacencies
are preserved. G1 and G2 are
both N-by-N sparse matrices that represent directed or undirected
graphs. Return value Isomorphic is Boolean.
When Isomorphic is true, Map is
a row vector containing the node indices that map from G2 to G1 for
one possible isomorphism. When Isomorphic is false, Map is
empty. The worst-case time complexity is O(N!),
where N is the number of nodes.
[
indicates whether the graphs are directed or undirected. Set Isomorphic, Map]
= graphisomorphism(G1, G2,'Directed', DirectedValue)DirectedValue to false when
both G1 and G2 are
undirected graphs. In this case, the upper triangles of the sparse
matrices G1 and G2 are
ignored. Default is true, meaning that both graphs
are directed.
Examples
Create and view a directed graph with 8 nodes and 11 edges.
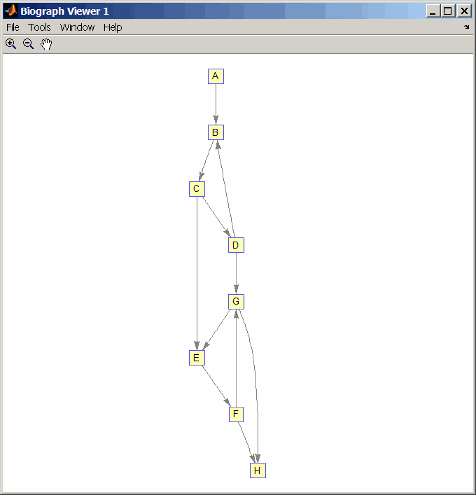
m('ABCDEFGH') = [1 2 3 4 5 6 7 8]; g1 = sparse(m('ABDCDCGEFFG'),m('BCBDGEEFHGH'),true,8,8) g1 = (1,2) 1 (4,2) 1 (2,3) 1 (3,4) 1 (3,5) 1 (7,5) 1 (5,6) 1 (4,7) 1 (6,7) 1 (6,8) 1 (7,8) 1 view(biograph(g1,'ABCDEFGH'))
Set a random permutation vector and then create and view a new permuted graph.
p = randperm(8) p = 7 8 2 3 6 4 1 5 g2 = g1(p,p); view(biograph(g2,'12345678'))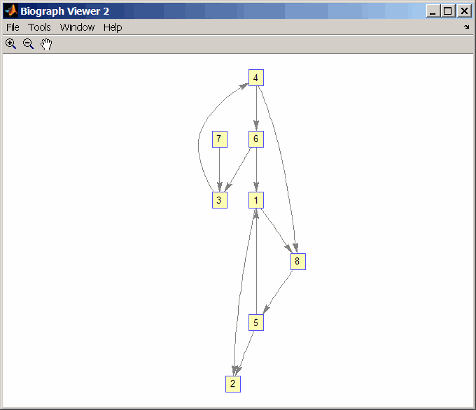
Check if the two graphs are isomorphic.
[F,Map] = graphisomorphism(g2,g1) F = 1 Map = 7 8 2 3 6 4 1 5Note that the
Maprow vector containing the node indices that map fromg2tog1is the same as the permutation vector you created in step 2.Reverse the direction of the D-G edge in the first graph, and then check for isomorphism again.
g1(m('DG'),m('GD')) = g1(m('GD'),m('DG')); view(biograph(g1,'ABCDEFGH'))
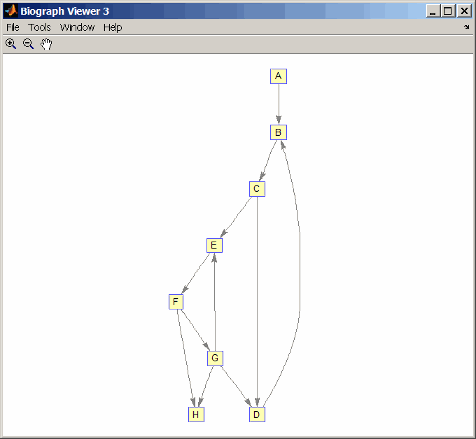
[F,M] = graphisomorphism(g2,g1) F = 0 M = []Convert the graphs to undirected graphs, and then check for isomorphism.
[F,M] = graphisomorphism(g2+g2',g1+g1','directed',false) F = 1 M = 7 8 2 3 6 4 1 5
References
[1] Fortin, S. (1996). The Graph Isomorphism Problem. Technical Report, 96-20, Dept. of Computer Science, University of Alberta, Edomonton, Alberta, Canada.
[2] McKay, B.D. (1981). Practical Graph Isomorphism. Congressus Numerantium 30, 45-87.
[3] Siek, J.G., Lee, L-Q, and Lumsdaine, A. (2002). The Boost Graph Library User Guide and Reference Manual, (Upper Saddle River, NJ:Pearson Education).
See Also
graphallshortestpaths | graphconncomp | graphisdag | graphisspantree | graphmaxflow | graphminspantree | graphpred2path | graphshortestpath | graphtopoorder | graphtraverse | isomorphism