graphallshortestpaths
Find all shortest paths in graph
Syntax
[dist] =
graphallshortestpaths(G)
[dist] =
graphallshortestpaths(G, ...'Directed', DirectedValue,
...)
[dist] = graphallshortestpaths(G,
...'Weights', WeightsValue, ...)
Arguments
G | N-by-N sparse matrix that represents a graph. Nonzero entries
in matrix G represent the weights of the
edges. |
DirectedValue | Property that indicates whether the graph is directed or undirected.
Enter false for an undirected graph. This results
in the upper triangle of the sparse matrix being ignored. Default
is true. |
WeightsValue | Column vector that specifies custom weights for the edges in
matrix G. It must have one entry for every
nonzero value (edge) in matrix G. The order
of the custom weights in the vector must match the order of the nonzero
values in matrix G when it is traversed
column-wise. This property lets you use zero-valued weights. By default, graphallshortestpaths gets
weight information from the nonzero entries in matrix G. |
Description
Tip
For introductory information on graph theory functions, see Graph Theory Functions.
[ finds
the shortest paths between every pair of nodes in the graph represented
by matrix dist] =
graphallshortestpaths(G)G, using Johnson's algorithm.
Input G is an N-by-N sparse matrix that
represents a graph. Nonzero entries in matrix G represent
the weights of the edges.
Output dist is an N-by-N matrix where dist(S,T)S to
target node T. Elements in the diagonal of this
matrix are always 0, indicating the source node
and target node are the same. A 0 not in the diagonal
indicates that the distance between the source node and target node
is 0. An Inf indicates there
is no path between the source node and the target node.
Johnson's algorithm has a time complexity of O(N*log(N)+N*E),
where N and E are the number
of nodes and edges respectively.
[...] = graphallshortestpaths ( calls G,
'PropertyName', PropertyValue,
...)graphallshortestpaths with
optional properties that use property name/property value pairs. You
can specify one or more properties in any order. Each PropertyName must
be enclosed in single quotes and is case insensitive. These property
name/property value pairs are as follows:
[ indicates whether the graph is directed
or undirected. Set dist] =
graphallshortestpaths(G, ...'Directed', DirectedValue,
...)DirectedValue to false for
an undirected graph. This results in the upper triangle of the sparse
matrix being ignored. Default is true.
[ lets
you specify custom weights for the edges. dist] = graphallshortestpaths(G,
...'Weights', WeightsValue, ...)WeightsValue is
a column vector having one entry for every nonzero value (edge) in
matrix G. The order of the custom weights
in the vector must match the order of the nonzero values in matrix G when
it is traversed column-wise. This property lets you use zero-valued
weights. By default, graphallshortestpaths gets
weight information from the nonzero entries in matrix G.
Examples
Create and view a directed graph with 6 nodes and 11 edges.
W = [.41 .99 .51 .32 .15 .45 .38 .32 .36 .29 .21]; DG = sparse([6 1 2 2 3 4 4 5 5 6 1],[2 6 3 5 4 1 6 3 4 3 5],W) DG = (4,1) 0.4500 (6,2) 0.4100 (2,3) 0.5100 (5,3) 0.3200 (6,3) 0.2900 (3,4) 0.1500 (5,4) 0.3600 (1,5) 0.2100 (2,5) 0.3200 (1,6) 0.9900 (4,6) 0.3800 view(biograph(DG,[],'ShowWeights','on'))
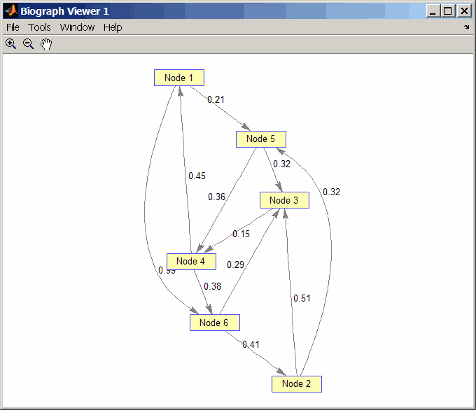
Find all the shortest paths between every pair of nodes in the directed graph.
graphallshortestpaths(DG) ans = 0 1.3600 0.5300 0.5700 0.2100 0.9500 1.1100 0 0.5100 0.6600 0.3200 1.0400 0.6000 0.9400 0 0.1500 0.8100 0.5300 0.4500 0.7900 0.6700 0 0.6600 0.3800 0.8100 1.1500 0.3200 0.3600 0 0.7400 0.8900 0.4100 0.2900 0.4400 0.7300 0The resulting matrix shows the shortest path from node 1 (first row) to node 6 (sixth column) is 0.95. You can see this in the graph by tracing the path from node 1 to node 5 to node 4 to node 6 (0.21 + 0.36 + 0.38 = 0.95).
Create and view an undirected graph with 6 nodes and 11 edges.
UG = tril(DG + DG') UG = (4,1) 0.4500 (5,1) 0.2100 (6,1) 0.9900 (3,2) 0.5100 (5,2) 0.3200 (6,2) 0.4100 (4,3) 0.1500 (5,3) 0.3200 (6,3) 0.2900 (5,4) 0.3600 (6,4) 0.3800 view(biograph(UG,[],'ShowArrows','off','ShowWeights','on'))

Find all the shortest paths between every pair of nodes in the undirected graph.
graphallshortestpaths(UG,'directed',false) ans = 0 0.5300 0.5300 0.4500 0.2100 0.8300 0.5300 0 0.5100 0.6600 0.3200 0.7000 0.5300 0.5100 0 0.1500 0.3200 0.5300 0.4500 0.6600 0.1500 0 0.3600 0.3800 0.2100 0.3200 0.3200 0.3600 0 0.7400 0.8300 0.7000 0.5300 0.3800 0.7400 0The resulting matrix is symmetrical because it represents an undirected graph. It shows the shortest path from node 1 (first row) to node 6 (sixth column) is 0.83. You can see this in the graph by tracing the path from node 1 to node 4 to node 6 (0.45 + 0. 38 = 0.83). Because
UGis an undirected graph, we can use the edge between node 1 and node 4, which we could not do in the directed graphDG.
References
[1] Johnson, D.B. (1977). Efficient algorithms for shortest paths in sparse networks. Journal of the ACM 24(1), 1-13.
[2] Siek, J.G., Lee, L-Q, and Lumsdaine, A. (2002). The Boost Graph Library User Guide and Reference Manual, (Upper Saddle River, NJ:Pearson Education).
See Also
allshortestpaths | graphconncomp | graphisdag | graphisomorphism | graphisspantree | graphmaxflow | graphminspantree | graphpred2path | graphshortestpath | graphtopoorder | graphtraverse