1-D Multisignal Analysis
This section takes you through the features of 1-D multisignal wavelet analysis, compression and denoising using the Wavelet Toolbox™ software. The rationale for each topic is the same as in the 1-D single signal case.
The toolbox provides the following functions for multisignal analysis.
Analysis-Decomposition and Synthesis-Reconstruction Functions
Decomposition Structure Utilities
Function Name | Purpose |
|---|---|
Change multisignal 1-D decomposition coefficients | |
Multisignal 1-D decomposition energy repartition |
Compression and Denoising Functions
You can perform analyses from the MATLAB® command line or by using the Wavelet Analyzer app. This section describes each method. The last section discusses how to exchange signal and coefficient information between the disk and the graphical tools.
1-D Multisignal Analysis — Command Line
Load a file, from the MATLAB prompt, by typing
load thinker
The file
thinker.matcontains a single variableX. Usewhosto show information aboutX.whos
Name Size Bytes Class X192x96147456double arrayPlot some signals.
figure; plot(X(1:5,:)','r'); hold on plot(X(21:25,:)','b'); plot(X(31:35,:)','g') set(gca,'Xlim',[1,96]) grid

Perform a wavelet decomposition of signals at level 2 of row signals using the
db2wavelet.dec = mdwtdec('r',X,2,'db2')This generates the decomposition structure
dec:dec = dirDec: 'r' level: 2 wname: 'db2' dwtFilters: [1x1 struct] dwtEXTM: 'sym' dwtShift: 0 dataSize: [192 96] ca: [192x26 double] cd: {[192x49 double] [192x26 double]}Change wavelet coefficients.
For each signal change the wavelet coefficients by setting all the coefficients of the detail of level 1 to zero.
decBIS = chgwdeccfs(dec,'cd',0,1);
This generates a new decomposition structure
decBIS.Perform a wavelet reconstruction of signals and plot some of the new signals.
Xbis = mdwtrec(decBIS); figure; plot(Xbis(1:5,:)','r'); hold on plot(Xbis(21:25,:)','b'); plot(Xbis(31:35,:)','g') grid; set(gca,'Xlim',[1,96])

Compare old and new signals by plotting them together.
figure; idxSIG = [1 31]; plot(X(idxSIG,:)','r','linewidth',2); hold on plot(Xbis(idxSIG,:)','b','linewidth',2); grid; set(gca,'Xlim',[1,96])

Set the wavelet coefficients at level 1 and 2 for signals 31 to 35 to the value zero, perform a wavelet reconstruction of signal 31, and compare some of the old and new signals.
decTER = chgwdeccfs(dec,'cd',0,1:2,31:35); Y = mdwtrec(decTER,'a',0,31); figure; plot(X([1 31],:)','r','linewidth',2); hold on plot([Xbis(1,:) ; Y]','b','linewidth',2); grid; set(gca,'Xlim',[1,96])

Compute the energy of signals and the percentage of energy for wavelet components.
[E,PEC,PECFS] = wdecenergy(dec);
Energy of signals 1 and 31:
Ener_1_31 = E([1 31]) Ener_1_31 = 1.0e+006 * 3.7534 2.2411Compute the percentage of energy for wavelet components of signals 1 and 31.
PEC_1_31 = PEC([1 31],:) PEC_1_31 = 99.7760 0.1718 0.0522 99.3850 0.2926 0.3225
The first column shows the percentage of energy for approximations at level 2. Columns 2 and 3 show the percentage of energy for details at level 2 and 1, respectively.
Display the percentage of energy for wavelet coefficients of signals 1 and 31. As we can see in the
decstructure, there are 26 coefficients for the approximation and the detail at level 2, and 49 coefficients for the detail at level 1.PECFS_1 = PECFS(1,:); PECFS_31 = PECFS(31,:); figure; plot(PECFS_1,'r','linewidth',2); hold on plot(PECFS_31,'b','linewidth',2); grid; set(gca,'Xlim',[1,size(PECFS,2)])

Compress the signals to obtain a percentage of zeros near 95% for the wavelet coefficients.
[XC,decCMP,THRESH] = mswcmp('cmp',dec,'N0_perf',95); [Ecmp,PECcmp,PECFScmp] = wdecenergy(decCMP);Plot the original signals 1 and 31, and the corresponding compressed signals.
figure; plot(X([1 31],:)','r','linewidth',2); hold on plot(XC([1 31],:)','b','linewidth',2); grid; set(gca,'Xlim',[1,96])

Compute thresholds, percentage of energy preserved and percentage of zeros associated with the
L2_perfmethod preserving at least 95% of energy.[THR_VAL,L2_Perf,N0_Perf] = mswcmptp(dec,'L2_perf',95); idxSIG = [1,31]; Thr = THR_VAL(idxSIG) Thr = 256.1914 158.6085 L2per = L2_Perf(idxSIG) L2per = 96.5488 94.7197 N0per = N0_Perf(idxSIG) N0per = 79.2079 86.1386
Compress the signals to obtain a percentage of zeros near 60% for the wavelet coefficients.
[XC,decCMP,THRESH] = mswcmp('cmp',dec,'N0_perf',60);XCsignals are the compressed versions of the original signals in the row direction.Compress the
XCsignals in the column directionXX = mswcmp('cmpsig','c',XC,'db2',2,'N0_perf',60);Plot original signals
Xand the compressed signalsXXas images.figure; subplot(1,2,1); image(X) subplot(1,2,2); image(XX) colormap(pink(222))

Denoise the signals using the universal threshold:
[XD,decDEN,THRESH] = mswden('den',dec,'sqtwolog','sln'); figure; plot(X([1 31],:)','r','linewidth',2); hold on plot(XD([1 31],:)','b','linewidth',2); grid; set(gca,'Xlim',[1,96])
XDsignals are the denoised versions of the original signals in the row direction.Denoise the
XDsignals in column directionXX = mswden('densig','c',XD,'db2',2,'sqtwolog','sln');Plot original signals
Xand the denoised signalsXXas images.figure; subplot(1,2,1); image(X) subplot(1,2,2); image(XX) colormap(pink(222))

1-D Multisignal Analysis Using the Wavelet Analyzer App
In this section, we explore the same signal as in the previous section, but use the Wavelet Analyzer app to analyze it.
Start the Wavelet 1-D Multisignal Analysis Tool.
From the MATLAB prompt, type
waveletAnalyzer.The Wavelet Analyzer appears.

Click Multisignal Analysis 1-D to open the Wavelet 1-D Multisignal Analysis tool.

The tool is divided into five panes. Two of them are the same as in all Wavelet Toolbox app tools, the Command Frame on the right side of the figure and the Dynamic Visualization tool at the bottom. The Command Frame contains a special component found in all multisignal tools: the Selection of Data Sets pane which is used to manage two lists.
The three new panes are the Visualization of Selected Data pane, the Information on Selected Data pane, and the Selection of Data pane.
Load the signals.
At the MATLAB command prompt, type
In the Multisignal Analysis 1-D tool, select File > Import from Workspace > Import Signals. When the Import from Workspace dialog box appears, select theload thinker
Xvariable. Click OK to import the data matrix and display the first signal.
The Selection of Data pane contains a list of selectable signals. At the beginning, only the originally loaded signals are available. You can generate and add new signals to the list by decomposing, compressing, or denoising original signals.
Each row of the list displays the index of selectable signal (
Idx Sel), the index of original signal (Idx Sig) and three wavelet transform attributes describing the process used to obtain the selectable signal from the original one.View the signals and signal information.
With signal 1 highlighted, Shift-click the mouse on signal 3 to select signals 1, 2, and 3.
Ctrl-click the mouse on signals 7, 9, and 11. (The Select ALL button at the bottom of the Selection of Data pane selects all signals and the Clear button deselects all signals.)
The selected signals (1, 2, 3, 7, 9 and 11) appear in the Visualization of Selected Data pane. The Information on Selected Data pane contains the box plots of the minimums, the means, and the maximums of these signals.

Highlight a signal.
Using the Highlight Sel button in the lower-left corner of the Visualization of Selected Data pane, select signal 3.

Select Different Views.
In the Visualization of Selected Data pane, change the view mode using the pop-up in the lower-right corner. Choose Separate Mode. The selected signals appear.
Decompose a multisignal.
Perform an analysis at level 4 using the
db2wavelet and the same file used in the command line section:thinker.mat.In the upper right portion of the Wavelet 1-D Multisignal Analysis tool, select
db2and level 4 in the Wavelet fields.
Click Decompose. After a pause for computation, all the original signals are decomposed and signal 1 is automatically selected

In the Selection of Data pane, new information is added for each original signal — the percentage of energy of the wavelet components (D1,...,D4 and A4) and the total energy. The Information on Selected Data pane contains information on the single selected signal: Min, Mean, Max and the energy distribution of the signal.

Since the original signals are decomposed, new objects appear and the Selection of Data Sets pane in the Command Frame updates.
The Selection of Data Sets pane defines the available signals that are now selectable from the Selection of Data pane.

The list on the left allows you to select sets of signals and the right list allows you to select sets of corresponding coefficients: original signals (
Orig. Signals), approximations (APP 1,...)and details from levels 1 to 4 (DET 1,...).In the list on the right, the coefficients vectors can be of different lengths, but only components of the same length can be selected together.
After a decomposition the original signals (
Orig. Signals) data set appears automatically selected.Select signals 1, 2, 3, 7, 9 and 11.

The energy of selected signals is primarily concentrated in the approximation
A4, so the box plot is crushed (see following figure on the left). Deselect App. On/Off to see a better representation of details energy (see following figure on the right).
Display multisignal decompositions.
In the Visualization of Selected Data pane, change the view mode using the pop-up below the plots and select
Full Dec Mode. The decompositions of the selected signals display.
Change the Level to
2.Select the signal 7 in Highlight Sel.
Change the visualization modes.
Using the second pop-up from the left at the bottom of the pane, select
Full Dec Mode (Cfs). The coefficients of the decompositions of the selected signals display. At level k, coefficients are duplicated 2k times.Change the view mode to
Stem Mode (Abs), and then change toTree Mode. The wavelet tree corresponding to the decompositions of the selected signals displays.Select the level 4 and click the node a3. Then highlight signal 7.
Select Different Wavelet Components.
Ctrl-click Orig. Signals, APP 1, APP 3 and DET 1 to select these four sets of signals from the list on the left in the Selection of Data Sets pane.
The total number of selected data (
Number of Sig.) appears in the Selection of Data Sets pane: four sets of 192 signals each is a total of 768 signals.
Click the Asc. button in the Sort pane. The selected data are sorted in ascending order with respect to the
Idx Sigparameter
Note that DWT attributes of each selectable signal have been updated where a stands for approximation, d for detail and s for signal.
Click the
Idx Sel 1signal and then shift-click theIdx Sel 579signal.
Choose
Separate Mode.
Ctrl-click to select two sets of signals from the right-most list of the Selection of Data Sets pane:
APP 1andDET 1.
Note that in this list of coefficients sets, the selected vectors must be of same length, which means that you must select components of the same level.
Click the Asc. button in the Sort pane. The selected data are sorted in ascending order with respect to Idx Sig parameter.
Select the ten first signals.

Compress a multisignal.
The Wavelet Analyzer app features a compression option with automatic or manual thresholding.
Click Compress, located in the lower-right side of the window. This displays the Compression window.

Note
The tool always compresses all the original signals when you click the Compress button.
Before compressing, choose the particular strategy for computing the thresholds. Select the adapted parameters in the Select Compression Method frame. Then, apply this strategy to compute the thresholds according to the current method, either to the current selected signals by clicking the Selected button, or to all signals by clicking the ALL button. For this example, accept the defaults and click the ALL button.

The thresholds for each level (ThrD1 to ThrD4), the energy ratio (En. Rat.) and the sparsity ratio (NbZ Rat.) are displayed in the Selection of Data pane.
Click the Compress button at the bottom of the Thresholding pane. Now you can select new data sets: compressed Signals, the corresponding approximations, details and coefficients.
Press the Ctrl key and click the Compressed item in the left list of the Selection of Data Sets pane. The original signals and their compressed versions are selected (2 x 192 = 384 signals).
Click the Asc. button at the bottom of the Selection of Data pane to sort the signals using
Idx Signumber.With the mouse, select the first four signals. They correspond to the original signals 1, 2 and the corresponding compressed signals 193, 194.

Click the Close button to close the Compression window.
Denoise a multisignal.
The Wavelet Analyzer app offers a denoising option with either a predefined thresholding strategy or a manual thresholding method. Using this tool makes very easy to remove noise from many signals in one step.
Display the Denoising window by clicking the Denoise button located in the bottom part of the Command Frame on the right of the window.

A number of options are available for fine-tuning the denoising algorithm. For this example, accept the defaults:
softtype of thresholding,Fixed form thresholdmethod, andScaled white noiseas noise structure.Click the ALL button in the Thresholding pane. The threshold for each level (ThrD1, ..., ThrD4) computes and displays in the Selection of Data pane.

Then click the Denoise button at the bottom of the Thresholding pane.
Ctrl-click the Denoised item in the list on the left of the Selection of Data Sets pane. The original signals and the corresponding denoised ones are selected (2 x 192 = 384 signals).
Click the Asc. button at the bottom of the Selection of Data pane to sort the signals according to the Idx Sig parameter.
With the mouse, select the first four signals. They correspond to the original signals 1, 2 and the corresponding denoised signals 193, 194

Choose
Separate Mode.
To view residuals, Ctrl-click the
Orig. Signal, theDenoisedand theResidualsitems in the list on the left of the Selection of Data Sets pane. Original, denoised and residual signals are selected (3 x 192 = 576 signals).Click the Asc. button at the bottom of the Selection of Data pane to sort the signals using the
Idx Sigparameter.With the mouse, select the first six signals. They correspond to the original signals 1, 2, the corresponding denoised signals 193, 194 and the residuals 385, 386.
Then, choose
Separate Mode.
Click Close to close the denoising tool. Then, click the Yes button to update the synthesized signals.

Manual Threshold Tuning
Choose a method, select one or several signals in the Selection of Data pane using the mouse and keys. Then click the Selected button. You can select another group of signals using the same method. Press the Denoise button to denoise the selected signal(s).

You can also use manual threshold tuning. Click the Enable Manual Thresholding Tuning button.

The horizontal lines in the wavelet coefficient axes (
cd1, ...,cd4) can be dragged using the mouse. This may be done individually, by group or all together depending on the values in the Select Signal and Selected Level fields in the Manual Threshold Tuning pane.
In the Wavelet 1-D Multisignal Analysis Compression tool, you can use two methods for threshold tuning: the By level thresholding method which is used in the Wavelet 1-D Multisignal Analysis Denoising tool, and the Global thresholding method.

You can drag the vertical lines in the Energy and Nb. Zeros Performances axes using the mouse. This can be done individually or all together depending on the values of Select Signal in the Manual Threshold Tuning pane.

The threshold value, L2 performance, and number of zeros performance are updated in the corresponding edit buttons in the Manual Threshold Tuning pane.
Statistics on Signals
You can display various statistical parameters related to the signals and their components. From the Wavelet 1-D Multisignal Analysis tool, click the Statistics button. Then select the signal 1 in the Selection of Data Sets pane.

Select the signals 1, 2, 3, 7, 9 and 11 in the Selection of Data pane, and display the corresponding boxplots and correlation plots.

To display statistics on many wavelet components, in the Selection Data Sets pane, in the left column, select
Orig.Signals,APP 1,DET 1, Denoised andResidualssignals. Then chooseSeparate Mode, and click the Asc. button in the Sort pane. The selected data are sorted in ascending order with respect toIdx Sigparameter. In the Selection of Data pane, select data related to signal 1.
Clustering Signals
Note
To use clustering, you must have Statistics and Machine Learning Toolbox™ software installed. For more information about clustering, including measuring distances between objects and determining the proximity of objects to each other, see Hierarchical Clustering (Statistics and Machine Learning Toolbox).
Click the Clustering button located in the Command Frame, which is in the lower right of the Wavelet 1-D Multisignal Analysis window to open the Clustering tool.

You can cluster various type of signals and wavelet components: original, denoised or compressed, residuals, and approximations or details (reconstructed or coefficients). Similarly, there are several methods for constructing partitions of data.

Use the default parameters (
OriginalandSignalin Data to Cluster, and inAscending Hierarchical, euclidean, ward, and6in Clustering) and click the Compute Clusters button.A full dendrogram and a restricted dendrogram display in the Selection by Dendrogram pane. For each signal, the cluster number displays in the Selection of Data pane.

Select one cluster, several clusters, or a part of a cluster.
Click the
xticklabel 3at the bottom of the restricted dendrogram. The links of the third cluster blink in the full dendrogram and the 24 signals of this class display in the Visualization of Selected Data pane. You can see their numbers in the Selection of Data pane.Clicking the line in the restricted or in the full dendrogram lets you select one cluster, several linked clusters, or a part of a cluster. For a more accurate selection, use the
Dilate Xand theTranslate Xsliders under the full dendrogram. You can also use the Yscale button located above the full dendrogram. The corresponding signals display in the Visualization of Selected Data pane and in the list of the Selection of Data pane.
You can use the horizontal line in the full dendrogram to change the number of clusters. Use the left mouse button to drag the line up or down.

Use the Show Clusters button to examine the clusters of the current partition. You can display the mean (or the median) of each cluster, the global standard deviation and the pointwise standard deviation distance around the mean (or the median). Each plot title contains: the number of signals in the cluster (
Nb) and percent of total, the global standard deviation (D) of the cluster, and two indices of quality,Q1andQ2.The
Q1andQ2indices can be interpreted as measures of how well a cluster is concentrated in multidimensional space. The valuesminandmaxare the minimum and maximum values, respectively, of the absolute values of the signal samples in the cluster. If the global standard deviationDis small, thenQ1is close to 1 andQ2is close to 0. In this case, the cluster is considered well-concentrated. IfDis large, thenQ1is small andQ2is large. In this case, the cluster is considered more disperse. For example, in Cluster 3 the minimum and maximum values of the absolute values of the signal samples are 5 and 220, respectively. ThenQ1= 1 − 37.208/220 = 0.831 andQ2= 37.208/(220-5) = 0.173.
Click the Store Current Partition button below the Clustering pane to store the current partition for further comparisons. A default name is suggested. Note that the 1-D Wavelet Multisignal Analysis tool stores the partitions and they are not saved on the disk.

Partitions
Build and store several partitions (for example, partitions with signals, denoised signals approximations at level 1, 2 and 3, and denoised signals). Then, click the Open Partition Manager button below the Store Current Partition button. The Partitions Management pane appears. The names of all stored partitions are listed.

Now, you can show, clear, or save the partitions (individually, selected ones, or all together).
To display partitions, select the
Ori Signalsand theDen Signalspartitions, and click the Selected button next to the Show Partitions label.The clusters are almost the same, but it is difficult to see this on the Selected Partitions axis, due to the scaling difference. Press the Apply button to renumber the clusters (starting from the selected partition as basic numbering) to compare the two partitions.

Only three signals are not classified in the same cluster for the two considered partitions.
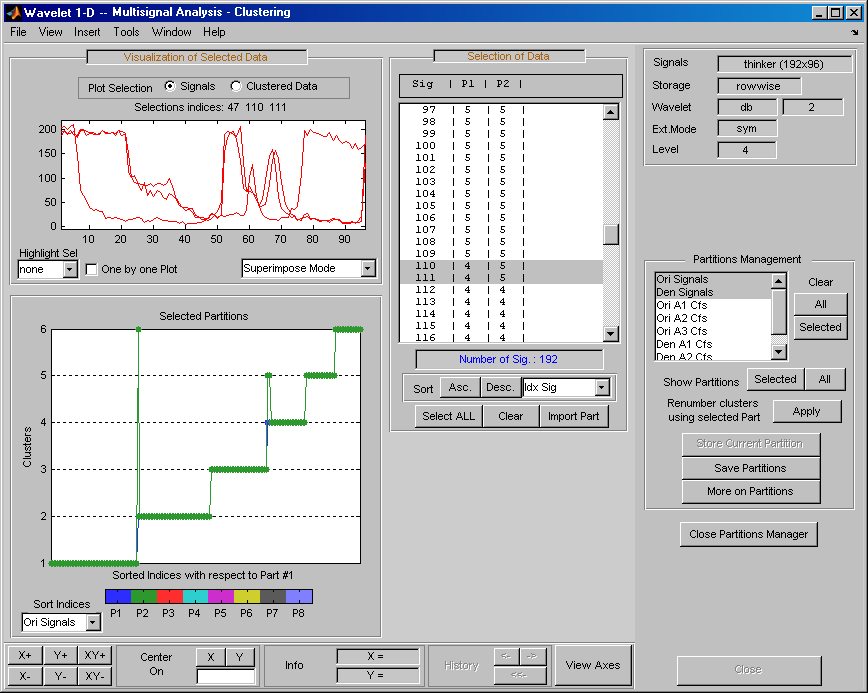
Select the partitions you want to save and click the Save Partitions button below the Store Current Partition button in the Partitions Management pane.

Partitions are saved as an array of integers, where each column corresponds to one partition and contains the indices of clusters. When you choose the
Full Partitionsoption, an array object (wpartobj) is saved.To load or clear stored partitions use File > Partitions in the Wavelet 1-D Multisignal Analysis tool. (File > Partitions is also available in the Wavelet 1-D Multisignal Analysis Clustering tool and you can also save the current partition.)

To clear one or more stored partitions, select File > Partitions > Clear Partition.

Select File > Partitions > Load Partition to load one or several partitions from the disk. The loaded partitions are stored in Wavelet 1-D Multisignal Analysis tool with any previously stored partitions. A partition can also be a manually created column vector.
Note
The number of signals in loaded partitions must be equal to the number of signals in the Wavelet 1-D Multisignal Analysis tool. A warning appears if this condition is not true.
In each subcomponent of the Wavelet 1-D Multisignal Analysis tool (main, statistics, denoising, compression, clustering), you can import a stored partition from the list in the Selection of Data pane. Click the Import Part button at the bottom of the Selection of Data pane, the Partition Set Manager window appears. Select one partition and click the Import button.

For this example, go back to the main window, import the
Ori Signalspartition and sort the signals in descending order with respect to A4 energy percentage.
You can compare partitions with the Partition tool. To display the Partition tool, click the More on Partitions button at the bottom of the Partitions Management pane. By default, when the Partition tool opens, the currently selected partition, in this case
Ori Signals, is compared with itself. In the lower left plot, an integerNat(i,j)means there is a group ofNsignals in theith cluster ofP1and thejth cluster ofP2. Since the Partition tool is comparing a partition with itself, all the numbers are plotted on the main diagonal.
Statistics measuring the similarity of the partitions
P1andP2are displayed in the panels on the right. The Partitions Pairs Links panel counts pairs of signals. Two signalsxandyare considered a pair if they are in the same cluster. For any two signalsxandy, there are four possibilities.The signals are paired in
P1andP2. This is called a true positive.The signals are paired in
P1but not inP2. This is called a false positive.The signals are not paired in
P1but are inP2. This is called a false negative.The signals are not paired in either
P1orP2. This is called a true negative.
The Partitions Pairs Links shows the percentages of each of the four possibilities. In this example, since there are 192 signals, there are total of 192*191/2 = 18336 possible pairs. Slightly more than 19% of the linked pairs are considered true positives, and approximately 80% are true negatives. Since the partition is being compared with itself, no pair is considered a false positive or false negative.
You can also measure how similar two partitions are by assigning a distance (or index) between them [1]. The distance is based on the number of true and false positives and negatives. Distance can be defined in different ways. Let R be the number of true positives, S be the number of true negatives, U be the number of false positives, and V be the number of false negatives. The Partitions Similarity Indices panel shows the distance between the
P1andP2partitions using different definitions of distance:Rand Index (
Rand): (R+S)/(R+S+U+V).Asymmetric Rand Index (
RandAsym): (2 × (R+S+U)+NS)/NS2 where NS is the total number of signals.Jaccard Index (
Jaccard): R/(R+U+V).Corrected Rand Index (
HubAra): (R-ER)/(MR-ER) where ER is the expected value of R and MR is the maximum value of the index. If the two partitions are identical, the index is equal to 1. It is possible for this index to be negative.Wallace Index (
Wallace): R/sqrt(|pi(P1)| × |pi(P2)|) where |pi(Pi)| is the number of joined pairs in the partitionPi.McNemar Index (
MacNemar): If abs(U+V) = 0, then the index is equal to 1. Otherwise, the index is equal to abs(U-V)/(U+V).Lerman Index (
ICL): (R-ER)/sqrt(VR) where VR is the variance of R.Normalized Lerman Index (
ILN):ICL(P1,P2)/sqrt(ICL(P1,P1) × ICL(P2,P2) where ICL(P,Q) denotes the Lerman index of two partitions P and Q.
Select the
Den Signalsin Sel P2 in the upper-right corner of the window. Then, in the lower left axis, click the yellow text containing the value2(the coordinates of the corresponding point are (4,5)). The corresponding signals are displayed together with all related information.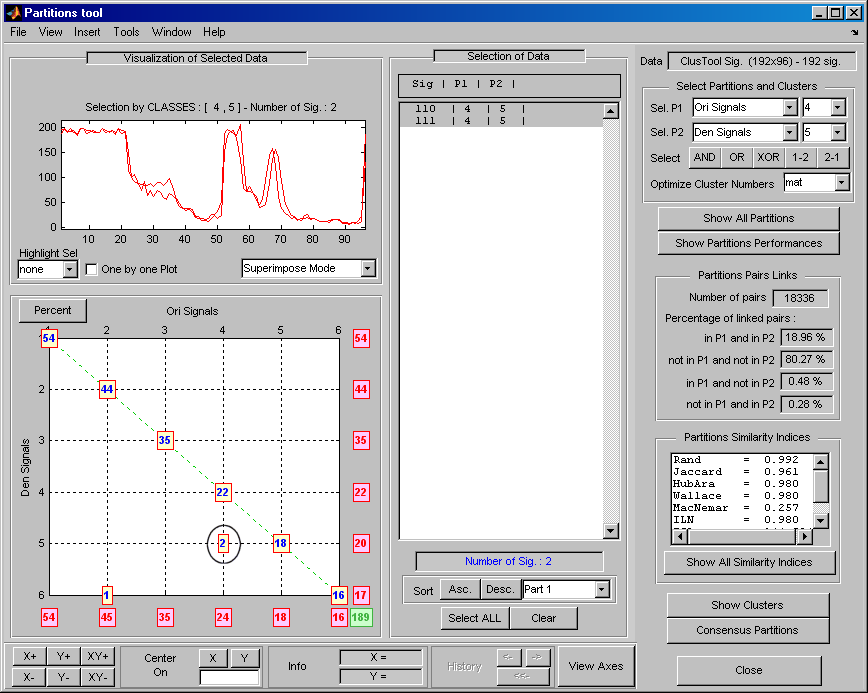
More on Clustering
Instead of the Ascending Hierarchical Tree clustering method, you can use the K-means method. For this case, the partition cannot be represented by a dendrogram and other representations should be used.
In the image representation (see figure below on the left), you can select a cluster by clicking on the corresponding color on the colorbar. You can also select a cluster or part of a cluster by clicking on the image.
In the center representation (see figure below on the right) you can select a cluster by clicking on the corresponding colored center.

Importing and Exporting Information from the Wavelet Analyzer App
The Wavelet 1-D Multisignal Analysis tool lets you move data to and from disk.
Saving Information to Disk
You can save decompositions and denoised or compressed signals (including the corresponding decompositions from Wavelet 1-D Multisignal Analysis tools) to disk. You then can manipulate the data and later import it again into the graphical tools.


Saving Decompositions
The Wavelet 1-D Multisignal Analysis main tool lets you save the entire set of data from a wavelet analysis to disk. The toolbox creates a MAT-file in the current folder with a name you choose.
Open the Wavelet 1-D Multisignal Analysis main tool and load the example analysis by selecting File > Example > Ex 21: Thinker (rows).
Save the data from this analysis, using the menu option File > Save Decompositions.
A dialog box appears that lets you specify a folder and filename for storing the decomposition data. For this example, use the name
decORI.mat.Type the name
decORI.After saving the decomposition data to the file
decORI.mat, load the variables into your workspace:load decORI whos
Name Size Bytes Class dec1x1163306structdec dec = dirDec: 'r' level: 4 wname: 'db2' dwtFilters: [1x1 struct] dwtEXTM: 'sym' dwtShift: 0 dataSize: [192 96] ca: [192x8 double] cd: {1x4 cell}The field
caof the structuredecgives the coefficients of approximation at level 4, the fieldcdis a cell array which contains the coefficients of details.size(dec.cd{1}) ans = 192 49 size(dec.cd{2}) ans = 192 26 size(dec.cd{3}) ans = 192 14 size(dec.cd{4}) ans = 192 8You can change the coefficients using the
chgwdeccfsfunction.Note
For a complete description of the
decstructure, see Loading Decompositions.
Loading Information into the Wavelet 1-D Multisignal Analysis Tool
You can load signals or decompositions into the graphical interface. The information you load may be previously exported from the graphical interface, and then manipulated in the workspace; or it may be information you initially generated from the command line. In either case, you must observe the strict file formats and data structures used by the Wavelet 1-D Multisignal Analysis tools or errors will occur when you try to load information.

Loading Signals. To load a signal you constructed in your MATLAB workspace
into the Wavelet 1-D Multisignal Analysis tool, save the signal in
a MAT-file (with extension .mat).
For example, if you design a signal called magic128 and
want to analyze it in the Wavelet 1-D Multisignal Analysis tool, type
save magic128 magic128
Note
The workspace variable magic128 must be a
matrix and the number of rows and columns must be greater than 1.
sizmag = size(magic128)
sizmag =
128 128
To load this signal into the Wavelet 1-D Multisignal Analysis tool, use the File > Load Signal menu item. A dialog box appears in which you select the appropriate MAT-file to be loaded.
Note
When you load a matrix of signals from the disk, the name of
2-D variables are inspected in the following order: x, X, sigDATA,
and signals. Then, the 2-D variables encountered
in the file are inspected in alphabetical order.
Loading Decompositions. To load decompositions that you constructed in the MATLAB workspace
into the Wavelet 1-D Multisignal Analysis tool, save the signal in
a MAT-file (with extension mat).
For instance, if you design a signal called magic128 and
want to analyze it in the Wavelet 1-D Multisignal Analysis too, the
structure dec must have the following fields:
'dirDec' | Direction indicator with |
'level' | Level of DWT decomposition |
'wname' | Wavelet name |
'dwtFilters' | Structure with four fields: |
'dwtEXTM' | DWT extension mode (see |
'dwtShift' | DWT shift parameter (0 or 1) |
'dataSize' | Size of original matrix |
'ca' | Approximation coefficients at level |
'cd' | Cell array of detail coefficients, from 1 to |
The coefficients cA and cD{k},
for (k = 1 to dec.level), are
matrices and are stored row-wise if dec.dirDec is
equal to 'r' or column-wise if dec.dirDec is
equal to 'c'.
Note
The fields 'wname' and 'dwtFilters' have
to be compatible (see the wfilters function).
The sizes of cA and cD{k}, (for k
= 1 to dec.level) must be compatible
with the direction, the level of the decomposition, and the extension
mode.
Loading and Saving Partitions.
Loading. The Wavelet 1-D Multisignal Analysis main tool and clustering tool let you load a set of partitions from disk.

Saving Partitions. The Wavelet 1-D Multisignal Analysis clustering tool lets you save a set of partitions to disk.

For example:
Open the Wavelet 1-D Multisignal Analysis main tool and load the example analysis using File > Example > Ex 21: Thinker (rows).
Click the Clustering button. The Wavelet 1-D Multisignal Analysis Clustering window appears.
Click the Compute Clusters button, and then save the current partition using menu option File > Partitions > Save Current Partition. A dialog box appears that lets you specify the type of data to save.
Click the Save Curr. button.

Another dialog box appears that lets you specify a folder and filename for storing the partition data. Type the name
curPART.After saving the partition data to the file
curPART.mat, load the variables into your workspace:load curPART whos
Name Size Bytes Class tab_IdxCLU192x11536doubleYou can modify the array
tab_IdxCLUin the workspace, and save the partition data in a file. For example to create two new partitions with four and two clusters, type the following lines:tab_IdxCLU(:,2) = rem((1:192)',4) + 1; tab_IdxCLU(:,3) = double((1:192)'>96) + 1; save newpart tab_IdxCLU
Now you can use three partitions for the example Ex 21: Thinker (rows). Then, you can load the partitions stored in the file
newPART.matin the Wavelet 1-D Multisignal Analysis main tool and clustering tool.Note
A partition is a column vector of integers. The values must vary from
1toNbClusters(NbClusters > 1), and each cluster must contain at least one element. The number of rows must be equal to the number of signals.
References
[1] Denoeud, L., Garreta, H., and A. Guénoche. "Comparison of Distance Indices Between Partitions." In International Symposium on Applied Stochastic Models and Data Analysis, 432–440. Brest, France: École Nationale des Télécommunications de Bretagne, 2005.