showalignment
(To be removed) Display color-coded sequence alignment
showalignment will be removed in a future release.
Syntax
showalignment(Alignment)
showalignment(..., 'MatchColor', MatchColorValue,
...)
showalignment(..., 'SimilarColor' SimilarColorValue,
...)
showalignment(..., 'StartPointers', StartPointersValue,
...)
showalignment(..., 'Columns', ColumnsValue,
...)
showalignment(..., 'TerminalGap', TerminalGapValue,
...)
Description
showalignment( displays a
color-coded sequence alignment in a MATLAB® Figure window. Alignment)
showalignment(..., ' calls
PropertyName',
PropertyValue, ...)showalignment with optional properties that use property
name/property value pairs. You can specify one or more properties in any order. Enclose
each PropertyName in single quotation marks. Each
PropertyName is case insensitive. These property
name/property value pairs are as follows:
showalignment(..., 'MatchColor',
specifies the color to highlight matching characters in the output display. MatchColorValue,
...)
showalignment(..., 'SimilarColor'
specifies the color to highlight similar characters in the output display. SimilarColorValue,
...)
showalignment(..., 'StartPointers',
specifies the starting indices in the original sequences of a local pairwise alignment. StartPointersValue,
...)
showalignment(..., 'Columns',
specifies the number of characters to display in one row when displaying a pairwise
alignment, and labels the start of each row with the sequence positions. ColumnsValue,
...)
showalignment(..., 'TerminalGap',
controls the inclusion or exclusion of terminal gaps from the count of matches and
similar residues when displaying a pairwise alignment.
TerminalGapValue,
...)TerminalGapValue can be true (default)
or false.
Input Arguments
|
Pairwise or multiple sequence alignment specified by one of the following:
|
|
Color to highlight matching characters in the output display. Specify the color with one of the following:
For example, to use cyan, enter Default: Red, which is specified by |
|
Color to highlight similar characters in the output display. Specify the color with one of the following:
For example, to use cyan, enter Default: Magenta, which is specified by |
|
Two-element vector that specifies the starting indices in the original sequences of a local pairwise alignment. Tip You can use the third output returned by |
|
Scalar that specifies the number of characters to display in one row when displaying a pairwise alignment. Default: |
|
Specifies whether to include or exclude terminal gaps from the count of
matches and similar residues when displaying a pairwise alignment. Choices
are |
Examples
Display a pairwise sequence alignment:
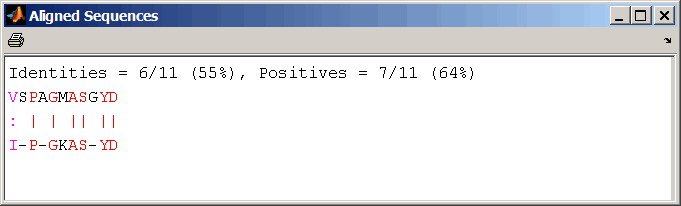
Notice that for pairwise sequence alignments, matching and similar characters appear in red and magenta respectively..
Display a multiple sequence alignment

Notice that for multiple sequence alignments, highly conserved positions appear in red and conserved positions appear in magenta.
Tip
To view a multiple-sequence alignment and interact with it, use the
seqalignviewer
function.
More About
Alternatives
You can also display a multiple or pairwise sequence alignment using the seqalignviewer function. The alignment displays in the Biological
Sequence Alignment window, where you can view and interactively adjust a sequence
alignment.
Compatibility Considerations
See Also
blosum | dayhoff | fastaread | gethmmalignment | gonnet | localalign | multialign | multialignread | nuc44 | nwalign | pam | seqalignviewer | showalignment | swalign