weights (phytree)
Calculate weights for phylogenetic tree
Syntax
W = weights(Tree)
Description
W = weights(Tree)Tree)
using the Thompson-Higgins-Gibson method. The distance of every segment
of the tree is adjusted by dividing it by the number of leaves it
contains. The sequence weights are the result of normalizing to unity
the new patristic distances between every leaf and the root.
Examples
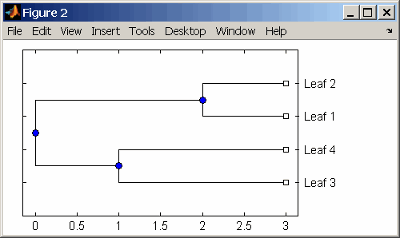
Create an ultrametric tree with specified branch distances.
bd = [1 2 3]'; tr_1 = phytree([1 2;3 4;5 6],bd)
View the tree.
view(tr_1)
Display the calculated weights.
weights(tr_1) ans = 1.0000 1.0000 0.8000 0.8000
References
[1] Thompson JD, Higgins DG, Gibson TJ (1994), "CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice," Nucleic Acids Research, 22(22):4673-4680.
[2] Henikoff S, Henikoff JG (1994), “Position-based sequence weights,” Journal Molecular Biology, 243(4):574-578.